Scientists spy on superbugs to see how they outsmart our antibiotics
Scientists have discovered yet another way that single-celled organisms have outsmarted us.
The tiny bacteria that live inside our guts have an ingenious way of withstanding the onslaught of antibiotics we throw at them, according to a report published Thursday in the journal Science. The two-part system allows bacterial cells to stay alive until another bacterium can deliver a lifeline, packaged in a snippet of DNA.
Microbes 1, humans 0.
“I’m afraid our findings are great news for bacterial cells — not so good for us,” said study leader Christian Lesterlin, a researcher in the Molecular Microbiology and Structural Biochemistry program at the University of Lyon in France.
Lesterlin and his colleagues already knew that superbugs could repel even our most modern medicines. What they didn’t know was how the microbes managed to pull it off.
“These are amazing abilities they have, to be able to adapt and survive in harsh environments with antibiotics,” he said. “But the more we understand about it, the more we can do for human health.”
READ MORE: A ‘slow catastrophe’ unfolds as the golden age of antibiotics comes to an end »
For most of human history, bacteria have had their way with us. Though some of them are helpful, others cause dangerous diseases like pneumonia, cholera and meningitis. The bacterium Yersinia pestis wiped out roughly 20% of the world’s population in the mid-1300s during the pandemic known as the Black Death.
When scientists first developed antibiotics in the early 1900s, humans enjoyed the upper hand — for a while. Some of the drugs target the machinery that maintains a bacterium’s all-important cell wall. Others rob bacteria of the proteins they need to carry out essential functions or damage the DNA needed to reproduce.
It took just a few decades for the first drug-resistant strains to appear. Since then, the invention of each new antibiotic invited a jeering reply.
Doctors responded by prescribing another antibiotic drug, and another. Then two drugs together. Then three. But now the arsenal is all but depleted, and there are strains of Escherichia coli, Klebsiella pneumoniae, Acinetobacter and Enterococcus that have evolved to overcome almost every medicine thrown at them.

So scientists are racing to understand superbugs’ tactics. Among the most urgent questions is this: How does antibiotic resistance spread between bacteria cells, even — or especially — in the presence of antibiotics that are designed to knock them back?
Bacteria know better than to wait around for a random mutation in their DNA that will protect them from antibiotics. Those mutations will come but not often: For some drugs, only about 1 in 10,000 bacteria will develop resistance that way. For other drugs, only about one in a billion will do so. Either way, that’s not very efficient.
Luckily for bacteria, they have plasmids at their disposal. These are circular snippets of DNA, and they can include genes that carry instructions for repelling specific antibiotics. Bacteria can swap useful plasmids with one another while socializing together in the human gut.
(Imagine having an anti-cancer gene and being able to pass out copies of it to everybody you bump into at the grocery store. Like I said, bacteria have outsmarted us.)
Lesterlin’s team wanted to visualize exactly how the exchange worked. They put a regular strain of Escherichia coli bacteria in one petri dish and a strain that is resistant to the antibiotic tetracycline in another dish. Then they saturated both plates with tetracycline and watched closely.
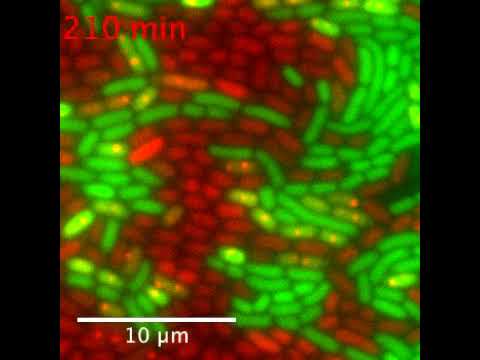
Logic suggested the bacteria cells lacking the ability to resist the drug would die. Instead, they simply went to sleep. After several hours, the researchers combined the contents of the two dishes and used a technique called live-cell microscopy to watch in real time as plasmids were transferred in just two minutes from tetracycline-resistant bacteria cells to tetracycline-sensitive ones.
Less than two hours later, the plasmid produced a protein called TetA resistance factor, which makes bacteria impervious to tetracycline. That was “shockingly counter-intuitive,” Lesterlin said, since tetracycline blocks the production of proteins by binding to the machinery required to make them.
The next question was this: How could bacteria get away with producing drug-resistance proteins right there in the presence of a protein-inhibiting drug?
As the hosts on QVC might say, one can never have too many accessories.
That’s especially true for something called the AcrAB-TolC multidrug efflux pump, which sits on the cell’s outer membrane and ejects various toxic antibiotics that have invaded the cell’s interior.
Despite the pump’s fancy-sounding name, it’s not sufficient to keep the cell thriving amid a surge of antibiotics. But it buys vital time for the groggy cell to acquire a plasmid with an all-important resistance gene.
In the team’s experiments, the pump kept tetracycline levels low enough to give the cell a chance to translate the resistance gene into a version of the TetA protein that was immune to the antibiotic.
Then that TetA protein took the reins in sustaining the newly drug-resistant cell, which went on to grow and multiply.
“Thanks to the multidrug efflux pump, bacteria have the ability to remain dormant — not quite dead but not quite alive — just waiting for a little help from a neighbor,” Lesterlin said.
Rudimentary microbiology discoveries like this one might seem far removed from the front lines of the war on superbugs. But military analysts and athletes alike will tell you that the key to victory is to study your enemy. Understanding basic cell functions could reveal gaping holes in our strategy to combat antibiotic resistance — and weaknesses of their own that we might exploit.
Lesterlin’s team has revealed “an essential piece of the puzzle,” said Vanessa Povolo, who studies bacterial growth strategies at the Swiss Federal Institute of Technology in Zurich and was not involved in the new work.
For one, the findings hint at the collateral damage from our century-old approach to fighting bacterial infections in humans. Overusing drugs has not only led to failed treatments but also has fueled the rise of newly resistant bacterial strains that come to life right in our digestive tracts.
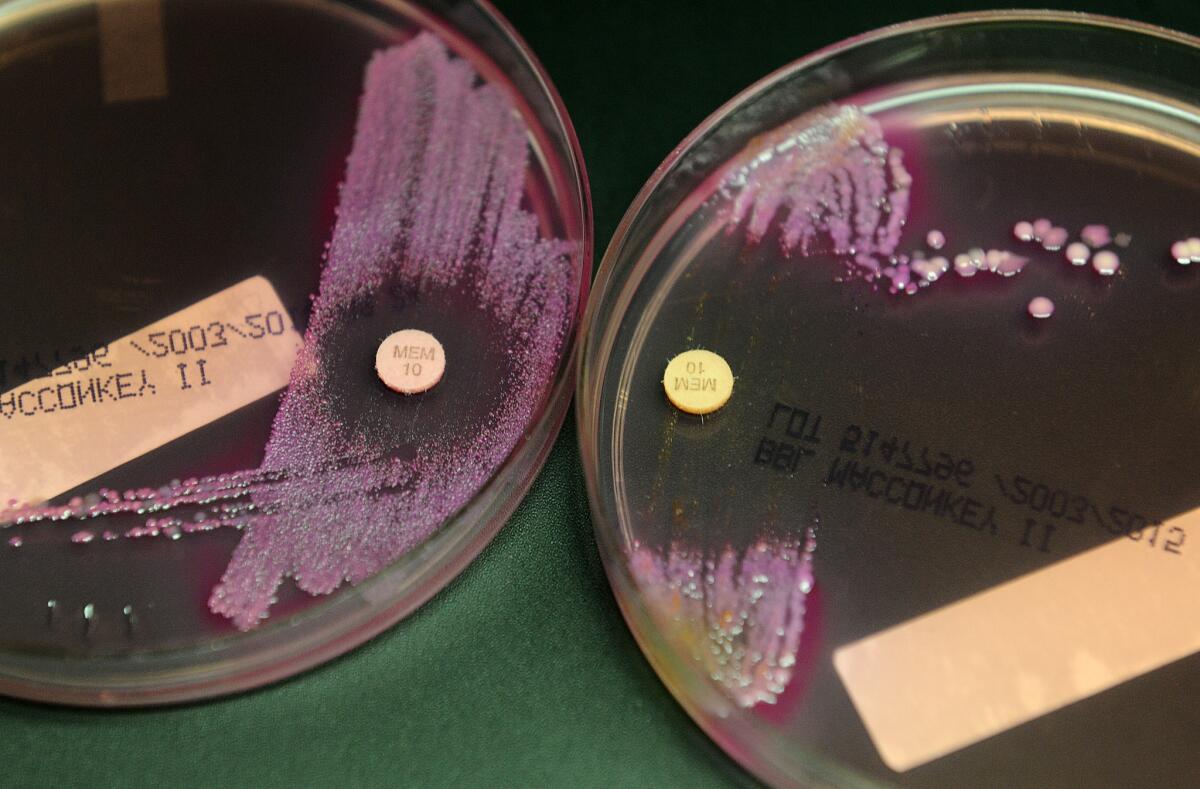
Now that scientists understand the mechanics of plasmid transfers, they can try to create new treatments that attack the multidrug efflux pumps that allow resistance to spread.
“Bacteria have multiple weapons — you can’t just shut down one weapon and expect to succeed,” said Shaun Yang, assistant medical director of the Clinical Microbiology Laboratory at UCLA, who was not involved in the research. “From a drug development perspective, that’s significant.”
For now, scientists remain locked in a race that could mean life or death for all organisms involved. Antibiotic-resistant bacteria already kill at least 23,000 people in the United States a year, according to the Centers for Disease Control and Prevention — an estimate that most experts consider conservative.
The United Nations warns that, without action, drug-resistant infections could kill 10 million people annually by 2050. That’s a public health nightmare, but hardly a surprise.
After all, bacteria have a few billion years of guerrilla warfare under their belts.